The CDCS version 3.X has a rest API that can be accessed via https://cdcs.nist.gov/docs/api#/rest. You can then click one of the operations like GET /rest/data. It will display that parameters and provide a sample for you to try. Below we will give sample Python 3.X code to access many of the operations. NOTE: You will need admin privledge to use ALL of the Rest API Calls. The examples provide the subroutines only and assume the user can create the main routine and pass required arguments.
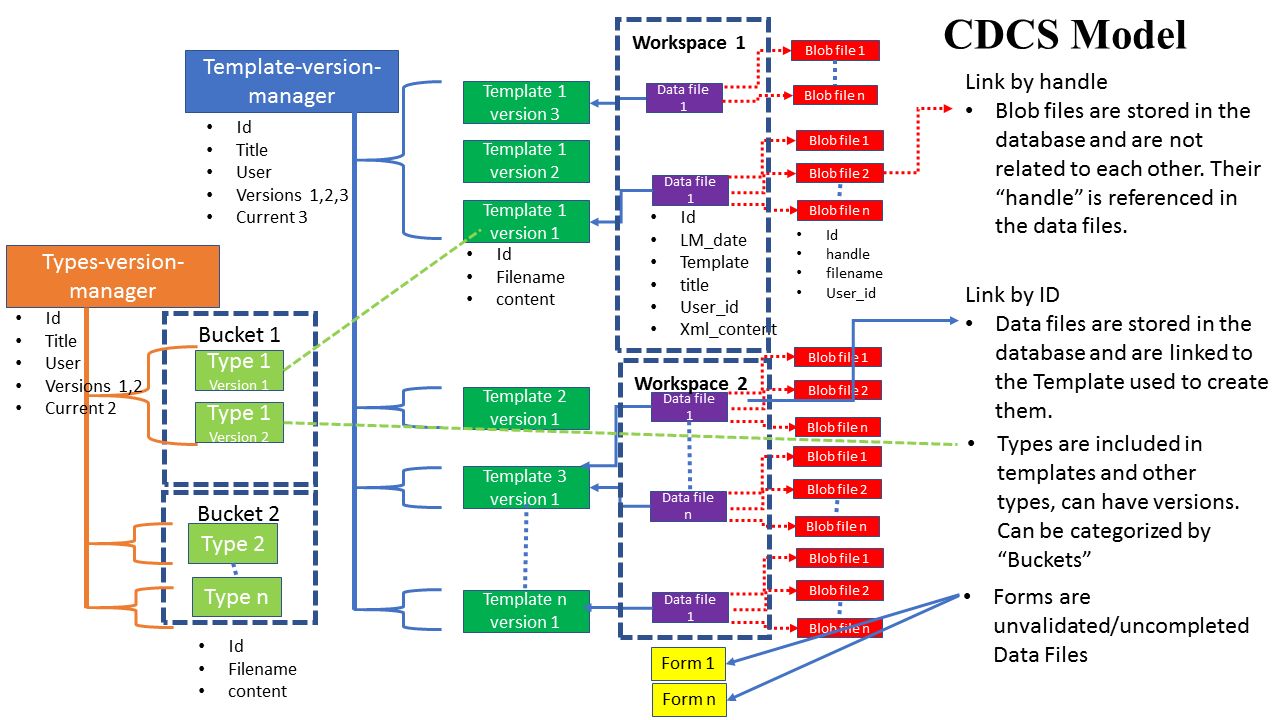
This is a pictoral representation of the model used by CDCS, XML Data files are created that validate to the associated schema definition in a template. TemplatesTemplates can have multiple versions, and are managed by the Template-version-manager. Binary "Blob" files representing any data via picture/spreadsheet can be included in an XML data files and referenced via a "handle" to the blob file. We will demonstrate accessing/created/deleting each of these in the following pages.
This will demonmstrate how to retrieve a template from the CDCS. You first call the Template-version-manager and it returns a list of all templates, each template will have the ID of the current version, and the ids of all previous versions. The code below will select all templates with the /rest/template-version-manager/global/ call, then select each templates info with the /rest/template/{current_id}/ call. A sample of the data values:
Template-verion-manager:
{u'_cls': u'VersionManager.TemplateVersionManager',
u'current': u'5c780edbd2d2051702a3510f',
u'disabled_versions': [],
u'id': u'5c780edbd2d2051702a35110',
u'is_disabled': False,
u'title': u'Potential',
u'user': None,
u'versions': [u'5c780edbd2d2051702a3510f']},
Template:
{u'content': u' (Content deleted for display) </xsd:element>\r\n</xsd:schema>',
u'dependencies': [],
u'filename': u'record-interatomic-potential.xsd',
u'hash': u'4b7dde5826186a44ef00d396265a02ef90d75382',
u'id': u'5c780edbd2d2051702a3510f'}
Note: Includes required in main program
import codecs
import json
import os
import re
import requests, zipfile, StringIO
import string
import subprocess
import sys
from glob import glob
from os import listdir
from os.path import join
from pprint import pprint
def get_all_templates(username='',pwd='',url=''):
"""
get_all_templates: get all templates from a curator
Parameters: username - username on CDCS instance (Note: User must have admin privileges)
password - password on CDCS instance
url - url of CDCS instance
"""
template_upload_url = "/rest/template-version-manager/global/"
turl = url + template_upload_url
print 'status: checking for lastes templates...'
response = requests.get(turl, verify=False, auth=(username, pwd))
print "Get:"
response_code = response.status_code
print "Response Code: "
print response_code
response_content = json.loads(response.text)
print "Template-version-manager:"
pprint(response_content)
print 'status: getting Templates...'
for rec in response_content:
cur=rec['current']
fetch_url = url + "/rest/template/" + cur + "/"
response = requests.get(fetch_url, verify=False, auth=(username, pwd))
out = response.json()
print "Template:"
pprint(out)
response_code = response.status_code
print "Response_code: "
print response.status_code
if response_code == requests.codes.ok:
print "status: Templates downloaded."
else:
response.raise_for_status()
raise Exception("- error: a problem occurred when uploading the schema (Error ", response_code, ")")
print 'status: done.'
OUTPUT:
status: checking for lastest templates...
Get:
Response Code:
200
status: getting Templates...
Template-version-manager:
[{u'_cls': u'VersionManager.TemplateVersionManager',
u'current': u'5c780edbd2d2051702a3510f',
u'disabled_versions': [],
u'id': u'5c780edbd2d2051702a35110',
u'is_disabled': False,
u'title': u'Potential',
u'user': None,
u'versions': [u'5c780edbd2d2051702a3510f']},
{u'_cls': u'VersionManager.TemplateVersionManager',
u'current': u'5c780edcd2d2051705a35111',
u'disabled_versions': [],
u'id': u'5c780edcd2d2051705a35112',
u'is_disabled': False,
u'title': u'Request',
u'user': None,
u'versions': [u'5c780edcd2d2051705a35111']},
.
. (output deleted for display)
Template:
{u'content': u'<?xml version="1.0" encoding="UTF-8" standalone="no"? (content removed for printing) ',
u'dependencies': [],
u'filename': u'record-interatomic-potential.xsd',
u'hash': u'4b7dde5826186a44ef00d396265a02ef90d75382',
u'id': u'5c06d071d2d2054b9d0fa1fb'}
Template:
{u'content': u'<?xml version="1.0" encoding="UTF-8" standalone="no"?> (content removed for printing) ',
u'dependencies': [],
u'filename': u'record-interatomic-potential-request.xsd',
u'hash': u'27d88e30f7af31b570967e3e37bf1fe4e7d1e29a',
u'id': u'5c06d083d2d2054b9b0fa2ff'}
Template:
{u'content': u'<?xml version="1.0" encoding="UTF-8" standalone="no"?> (content removed for printing) ',
u'dependencies': [],
u'filename': u'record-interatomic-potential-family.xsd',
u'hash': u'4bc28ba6f5a25afedd83582e089fe76dd1e73a53',
u'id': u'5c06d085d2d2054b9d0fa31d'}
Template:
{u'content': u'<?xml version="1.0" encoding="UTF-8" standalone="no"?>\r\n<xsd:schema xmlns:xsd="http://www.w3.org/2001/XMLSchema" attributeFormDef
ault="unqualified"\r\n elementFormDefault="unqualified">\r\n\r\n <xsd:element name="faq">\r\n <xsd:complexType>\r\n <xsd:s
equence>\r\n <xsd:element name="question" type="xsd:string"/>\r\n <xsd:element name="answer" type="xsd:string"/>\r\n
</xsd:sequence>\r\n </xsd:complexType>\r\n </xsd:element>\r\n</xsd:schema>',
u'dependencies': [],
u'filename': u'record-interatomic-potential-faq.xsd',
u'hash': u'12ba7d0270c5fa8d169ff54a5ef24e558e94b0d4',
u'id': u'5c06d086d2d2054b9e0fa323'}
Template:
{u'content': u'<?xml version="1.0" encoding="UTF-8" standalone="no"?> (content removed for printing) ',
u'dependencies': [],
u'filename': u'record-interatomic-potential-action.xsd',
u'hash': u'53b2f757bb148da3c461b65f1a61ff1080f3651c',
u'id': u'5c06d087d2d2054b9c0fa327'}
Template:
{u'content': u'<?xml version="1.0" encoding="UTF-8" standalone="no"?>\r\n<xsd:schema xmlns:xsd="http://www.w3.org/2001/XMLSchema" attributeFormDef
ault="unqualified"\r\n elementFormDefault="unqualified">\r\n\r\n <xsd:element name="per-potential-properties" type="xsd:anyType"/>\r\n</xsd:
schema>',
u'dependencies': [],
u'filename': u'record-per-potential-properties.xsd',
u'hash': u'1c54ec4f8ecd0b6600ae00846a6c1eb52d1b67ed',
u'id': u'5c06d091d2d2054b9e0fa3c9'}
.
.
.
for each template
.
.
Response_code:
200
status: Templates downloaded.
status: done.
(get the template named Diffraction_Data)
def get_template(username='',pwd='',url='', title=''):
"""
get_template: get a single template
parameters: username - username on CDCS instance (Note: User must have admin privledges)
password - password on CDCS instance
url - url of CDCS instance
title - Title of template to get
"""
template_upload_url = "/rest/template-version-manager/global/"
turl = url + template_upload_url
fetch_url = ''
print 'status: checking for lastes schema...'
response = requests.get(turl, verify=False, auth=(username, pwd))
print "Get:"
response_code = response.status_code
print "Response Code: "
print response_code
response_content = json.loads(response.text)
for rec in response_content:
print rec['title']
if ( title == rec['title']):
cur=rec['current']
fetch_url = url + "/rest/template/" + cur + "/"
if fetch_url == '':
response.raise_for_status()
raise Exception("Error: template not found (Error ", title, ")")
else :
turl = fetch_url
print 'status: getting Template...'
print "Get:"
response = requests.get(turl, verify=False, auth=(username, pwd))
out = response.json()
pprint(out)
response_code = response.status_code
print "Response_code: "
print response.status_code
if response_code == requests.codes.ok:
print "status: Templates downloaded."
else:
response.raise_for_status()
raise Exception("- error: a problem occurred when uploading the schema (Error ", response_code, ")")
print 'status: done.'
OUTPUT:
(get the template named Diffraction_Data)
status: getting Template...
Get:
{u'content': u'<?xml version="1.0" encoding="UTF-8" standalone="no"?>\r\n<xsd: ( rest of content delted for Display)
</xsd:complexType>\n</xsd:schema>',
u'dependencies': [],
u'filename': u'record-HTE-xray-diffraction.xsd',
u'hash': u'a4c73aa88525548d8da4cc132dbf5359d699a341',
u'id': u'5c780bb1d2d205146afb20f8'}
This picture will be used for data and blob file calls.
(this example gets the data file "faq")
def get_data_title(username='',pwd='',url='', data_file_title=''):
"""
get_data_title: get a single data file from curator by title
Parameters: username - username on CDCS instance (Note: User must have admin privledges)
password - password on CDCS instance
url - url of CDCS instance
data_file_title - get data file matching this title
"""
template_upload_url = "/rest/data"
turl = url + template_upload_url
print '- status: Getting Data file...'
data = {
"title":data_file_title
}
print "Get:"
response = requests.get(turl, params=data, verify=False, auth=(username, pwd))
out = response.json()
pprint(out)
print "Resp: "
print response.status_code
response_code = response.status_code
if response_code == requests.codes.ok:
print "- status: downloaded."
else:
response.raise_for_status()
raise Exception("- error: a problem occurred when uploading the schema (Error ", response_code, ")")
print '- status: done.'
OUTPUT:
(this example gets the data file "faq")
- status: Getting Data file...
Get:
[{u'id': u'5c06d086d2d2054b9c0fa321',
u'last_modification_date': u'2018-12-04T19:07:50.614000Z',
u'template': u'5c06d086d2d2054b9e0fa323',
u'title': u'faq',
u'user_id': u'1',
u'xml_content': u'<faq xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" ><question>What is the purpose of this project?</question><answer>The purpose of this project is to provide a repository of interatomic potentials for atomistic simulations (e.g. molecular dynamics) with comparison tools and reference experimental and ab-initio data in order to facilitate the evaluation of these potentials for particular applications. Our goal is not to judge that any particular potential is "the best" because the best interatomic potential may depend on the problem being considered. For example, some interatomic potentials which have been fit only to the properties of solid phases may model solid surface properties better than one fit with solid and liquid properties. However, the second potential will probably better represent properties which have a strong liquid contribution (e.g. crystal-melt interfacial properties). Additionally, we are not limiting the repository to a single class of material (e.g. metals), interatomic potential format (e.g. Embedded-Atom Method), or software package. As we obtain interatomic potentials for other materials or in other formats, we will include them with proper website modifications.</answer></faq>'}]
Resp:
200
- status: downloaded.
- status: done.
(This example looks up template by id - 5c06d072d2d2054b9b0fa1fd)
def get_data_full(username='',pwd='',url='', id=''):
"""
get_data_full: gat data file infor including it template's info
Parameters: username - username on CDCS instance (Note: User must have admin privledges)
password - password on CDCS instance
url - url of CDCS instance
id - id of data file to get info on
"""
template_upload_url = "/rest/data/get-full"
turl = url + template_upload_url
# upload the schema using REST
print 'status: Getting Data file info.'
data = {
"id":id
}
print "Get:"
response = requests.get(turl, params=data, verify=False, auth=(username, pwd))
out = response.json()
pprint(out)
print "Resp: "
print response.status_code
response_code = response.status_code
if response_code == requests.codes.ok:
print "- status: downloaded."
else:
response.raise_for_status()
raise Exception("- error: a problem occurred when uploading the schema (Error ", response_code, ")")
print '- status: done.'
OUTPUT:
(This example looks up template by id - 5c06d072d2d2054b9b0fa1fd)
status: Getting Data file info.
Get:
{u'id': u'5c06d072d2d2054b9b0fa1fd',
u'last_modification_date': u'2018-12-04T19:07:30.053000Z',
u'template': {u'_cls': u'Template',
u'_display_name': u'Potential (Version 1)',
u'content': u'<?xml version="1.0" encoding="UTF-8" standalone="no"?>\r\n<xsd:sche (Content removed for Diaply purposes) ',
u'dependencies': [],
u'filename': u'record-interatomic-potential.xsd',
u'hash': u'4b7dde5826186a44ef00d396265a02ef90d75382',
u'id': u'5c06d071d2d2054b9d0fa1fb'},
u'title': u'potential.2007--Munetoh-S-Motooka-T-Moriguchi-K-Shintani-A--Si-O',
u'user_id': u'1',
u'xml_content': u'<?xml version="1.0" encoding="utf-8"?>\n<interatomic-' (Content removed for Diaply purposes) }
Resp:
200
- status: downloaded.
- status: done.
This example uses - test.xml "5c06d071d2d2054b9d0fa1fb"
<filename) <Template ID)
def upload_data(username='',pwd='',url='',filen='',template_id=''):
"""
upload_data: import data file into a curator using specific template.
Parameters: username - username on CDCS instance (Note: User must have admin provledges)
password - password on CDCS instance
url - url of CDCS instance
filen - data file name to uplaod
template_id - id of template to use
"""
print 'status: uploading data file...'
xml_file = open(filen, 'rb');
xml_content = xml_file.read();
xml_upload_url = "/rest/data/"
turl = url + xml_upload_url
print "File:" + filen
print "Temp:" + template_id
data = {
"title": filen,
"template": template_id,
"xml_content": xml_content
}
response= requests.post(turl, data=data, auth=(username, pwd))
print "json:"
out = response.json();
pprint(out)
response_code = response.status_code
if response_code == HTTP_201_CREATED:
print "status: uploaded."
else:
response.raise_for_status()
raise Exception("- error: a problem occurred when uploading the schema (Error ", response_code, ")")
OUTPUT:
This example uses - test.xml "5c06d071d2d2054b9d0fa1fb"
(filename) (Template ID)
status: uploading data file...
File:test.xml
Temp:5c06d071d2d2054b9d0fa1fb
json:
{u'id': u'5c549316d2d20511cad31b5b',
u'last_modification_date': u'2019-02-01T18:42:30.467881Z',
u'template': u'5c06d071d2d2054b9d0fa1fb',
u'title': u'test.xml',
u'user_id': u'1',
u'xml_content': u'<?xml version="1.0" encoding="utf-8"?>\n<interatomic-potential><key>55d00f32-3bb5-433b-b01f-bef7c379d852</key><id>2007--Munetoh-S-Motooka-T-Moriguchi-K-Shintani-A--Si-O</id><record-version>2018-10-10</record-version><description><citation><document-type>journal</document-type><title>Interatomic potential for Si-O systems using Tersoff parameterization</title><author><given-name>S.</given-name><surname>Munetoh</surname></author><author><given-name>T.</given-name><surname>Motooka</surname></author><author><given-name>K.</given-name><surname>Moriguchi</surname></author><author><given-name>A.</given-name><surname>Shintani</surname></author><publication-name>Computational Materials Science</publication-name><publication-date><year>2007</year></publication-date><volume>39</volume><issue>2</issue><abstract>A parameter set for Tersoff potential has been developed to investigate the structural properties of Si-O systems. The potential parameters have been determined based on ab initio calculations of small molecules and the experimental data of \\\\u03b1-quartz. The structural properties of various silica polymorphs calculated by using the new potential were in good agreement with their experimental data and ab initio calculation results. Furthermore, we have prepared SiO<sub>2</sub> glass using molecular dynamics (MD) simulations by rapid quenching of melted SiO<sub>2</sub>. The radial distribution function and phonon density of states of SiO<sub>2</sub> glass generated by MD simulation were in excellent agreement with those of SiO<sub>2</sub> glass obtained experimentally.</abstract><pages>334-339</pages><DOI>10.1016/j.commatsci.2006.06.010</DOI><bibtex>@article{Munetoh_2007,\\\\n abstract = {A parameter set for Tersoff potential has been developed to investigate the structural properties of Si-O systems. The potential parameters have been determined based on ab initio calculations of small molecules and the experimental data of \\\\u03b1-quartz. The structural properties of various silica polymorphs calculated by using the new potential were in good agreement with their experimental data and ab initio calculation results. Furthermore, we have prepared SiO<sub>2</sub> glass using molecular dynamics (MD) simulations by rapid quenching of melted SiO<sub>2</sub>. The radial distribution function and phonon density of states of SiO<sub>2</sub> glass generated by MD simulation were in excellent agreement with those of SiO<sub>2</sub> glass obtained experimentally.},\\\\n author = {Shinji Munetoh and Teruaki Motooka and Koji Moriguchi and Akira Shintani},\\\\n doi = {10.1016/j.commatsci.2006.06.010},\\\\n journal = {Computational Materials Science},\\\\n month = {apr},\\\\n number = {2},\\\\n pages = {334--339},\\\\n publisher = {Elsevier BV},\\\\n title = {Interatomic potential for Si-O systems using Tersoff parameterization},\\\\n url = {https://doi.org/10.1016%2Fj.commatsci.2006.06.010},\\\\n volume = {39},\\\\n year = {2007}\\\\n}\\\\n\\\\n</bibtex></citation></description><implementation><key>08655b43-b844-466f-8e48-e908a9decbbb</key><id>2007--Munetoh-S--Si-O--LAMMPS--ipr1</id><status>active</status><date>2018-10-10</date><type>LAMMPS pair_style tersoff</type><notes><text>This file was created and verified by Lucas Hale. The parameter values are comparable to the SiO.tersoff file in the August 22, 2018 LAMMPS distribution, with this file having higher numerical precision for the derived mixing parameters.</text></notes><artifact><web-link><URL>https://www.ctcms.nist.gov/potentials/Download/2007--Munetoh-S-Motooka-T-Moriguchi-K-Shintani-A--Si-O/1/2007_SiO.tersoff</URL><link-text>2007_SiO.tersoff</link-text></web-link></artifact></implementation><element>Si</element><element>O</element></interatomic-potential>\n\n'}
status: uploaded.
This example uploads file test.fig
ddef upload_blob_file(username='',pwd='',url='',filen=''):
"""
upload_blob_file: import a single blob file into a curator.
Parameters: username - username on CDCS instance (Note: User must have admin privileges)
password - password on CDCS instance
url - url of CDCS instance
filen - name of blob file to load
"""
#
# If oid is pre-pended to filename remove it
#
oldid = filen[0:24]
if all(c in string.hexdigits for c in oldid):
newfile = filen[25:]
else:
newfile = filen
print '- status: uploading blob files ...'
xml_upload_url = "/rest/blob/"
xml_file = open(filen, 'rb');
xml_content = xml_file.read();
fd = open(newfile, 'wb');
fd.write(xml_content)
xml_file = {'blob': open(newfile, 'rb')}
turl = url + xml_upload_url
print url
print oldid
print newfile
print filen
data = {
"filename":newfile
}
print "Post:"
response= requests.post(turl, files=xml_file,data=data, auth=(username, pwd))
print "json:"
out = response.json();
response_code = response.status_code
print "Resp: "
print response
convert = "blob_convert/" + oldid
with open(convert, 'wb') as fd:
fd.write(str(out['handle']))
if response_code == requests.codes.created:
print "- status: uploaded."
os.remove(newfile);
else:
print "- error: Upload failed with status code "
print response_code
print ":"
print response["message"]
OUTPUT:
This example uploads file test.fig
status: uploading blob file...
json:
{u'filename': u'test.fig',
u'handle': u'http://vm-itl-ssd-017.nist.gov/rest/blob/download/5c549de1d2d20511c9d31b5f/',
u'id': u'5c549de1d2d20511c9d31b5f',
u'upload_date': u'2019-02-01 19:28:33+00:00',
u'user_id': u'1'}
status: uploaded.
This example downloads blob file with id=5c6da853d2d2054d15d2c434
def save_blob_to_file(username='',pwd='',url='',bid=''):
"""
save_blob_to_file: script to get a blob by id from a curator and save it to the original filename it was stored with.
Parameters: username - username on CDCS instance (Note: User must have admin privileges)
password - password on CDCS instance
url - url of CDCS instance
bid - id of the blod to extract, will create a local file using filename in CDCS of the blob
"""
print 'Get Blob id: ' + str(bid)
xml_upload_url = "/rest/blob/" + bid + "/"
turl = url + xml_upload_url
response = requests.get(turl, verify=False, auth=(username, pwd), stream=True)
out = response.json()
xml_upload_url = "/rest/blob/download/" + bid + "/"
turl = url + xml_upload_url
blobfile = requests.get(turl, verify=False, auth=(username, pwd), stream=True)
fname = out['filename']
with open(fname, 'wb') as fd:
fd.write(blobfile.content)
fd.close();
response_code = blobfile.status_code
if response_code == requests.codes.ok:
print "status: blob created: " + fname
ret = bid
else:
print "status: Error getting blob: " + response_code
ret = 0
return ret
OUTPUT:
This example downloads blob file with id=5c6da853d2d2054d15d2c434
Get Blob id: 5c6da853d2d2054d15d2c434
status: blob created: r95r88-1000h-labelled.tif
This example looks for data files with original filename "test"
def get_data_by_title(username='',pwd='',url='', data_file_title=''):
"""
Parameters: username - username on CDCS instance (Note: User must have admin privledges)
password - password on CDCS instance
url - url of CDCS instance
data_file_title - get data file matching this title
"""
template_upload_url = "/rest/data"
turl = url + template_upload_url
print '- status: Getting Data file...'
data = {
"title":data_file_title
}
print "Get:"
response = requests.get(turl, params=data, verify=False, auth=(username, pwd))
out = response.json()
pprint(out)
print "Resp: "
print response.status_code
response_code = response.status_code
if response_code == requests.codes.ok:
print "- status: downloaded."
else:
response.raise_for_status()
raise Exception("- error: a problem occurred when uploading the schema (Error ", response_code, ")")
print '- status: done.'
OUTPUT:
This example looks for data files with original filename "testing.txt"
status: Getting Data File ...testing.xml
Get:
[ {u'filename': u'testing.txt',
u'handle': u'http://vm-itl-ssd-017.nist.gov/rest/blob/download/5c0aa538d2d2054b9d0fa540/',
u'id': u'5c0aa538d2d2054b9d0fa540',
u'upload_date': u'2018-12-07 16:52:08+00:00',
u'user_id': u'5'}]
- status: blobs listed.
- status: done.
This example deletes a blob with id="5c549fa3d2d20511c7d31b5c"
def del_blob(username='',pwd='',url='',bid=''):
"""
del_blob: script to delete a blob data file from a curator.
Parameters: username - username on CDCS instance (Note: User must have admin privileges)
password - password on CDCS instance
url - url of CDCS instance
bid - id of blob to delete
"""
blob_delete_url = "/rest/blob/" + bid + "/"
turl = url + blob_delete_url
print 'status: Deleteing Blob file...' + bid
print "Delete: " + turl
del_response = requests.delete(turl, verify=False, auth=(username, pwd))
if del_response.status_code == 204:
print "- status: " + bid + " has been deleted."
else:
del_response.raise_for_status()
raise Exception("- error: a problem occurred when deleting the blobs (Error ", del_response.status_code, ")")
print 'status: done.'
OUTPUT:
This example deletes a blob with id="5c549fa3d2d20511c7d31b5c"
status: Deleteing Blob file...5c54a7e3d2d20511cbd31b5f
Delete: http://vm-itl-ssd-017.nist.gov:8050/rest/blob/5c54a7e3d2d20511cbd31b5f/
- status: 5c54a7e3d2d20511cbd31b5f has been deleted.
status: done.
def dump_curator(username='',pwd='',url=''):
"""
dump_curator: get all templates and data files and associated blobs from a 3.0 curator and dump to the local files system
Parameters: username - username on CDCS instance (Note: User must have admin priveledges)
password - password on CDCS instance
url - url of CDCS instance
creates a directory structure as so:
(Template Title)
(schema file.xsd)
(data files) (blob files)
(data_file1.xml) (blob_file1)
(data_file2.xml) (blob_file2)
(data_file3.xml) (blob_file3)
(data_file4.xml) (blob_file4)
(data_file5.xml) .
. .
. .
. .
(data_fileN.xml) (blob_fileX)
This strucure is created by this script . It will pre-pend the blobfile with it's id used in the data file to preserve relationship.
"""
template_url = "/rest/template-version-manager/global"
turl = url + template_url
print 'status: getting Templates...' + turl
response = requests.get(turl, verify=False, auth=(username, pwd))
out = json.loads(response.text)
return_code = subprocess.call("rm -rf ./schemas", shell=True)
return_code = subprocess.call("mkdir ./schemas", shell=True)
script = open("./loadata", "w+")
list1 = re.compile('http:\/\/127.0.0.1\/rest\/blob\/download\/(.*?)\<')
for rec in out:
ver=rec['versions']
title=rec['title']
current=rec['current']
print "Working on;" + title + " Current: " + current
counter=0
for templ in ver:
counter=counter+1
return_code = subprocess.call("mkdir ./schemas/" + title + str(counter), shell=True)
#pprint(templ)
template_url = "/rest/template/" + templ + "/"
turl = url + template_url
response1 = requests.get(turl, verify=False, auth=(username, pwd))
print response1.status_code
out1 = json.loads(response1.text)
if templ == current:
fn = "Cur_" + out1['filename'].encode('utf8')
else:
fn = out1['filename'].encode('utf8')
fff = open("./schemas/" + title + str(counter) +"/" + fn, "w")
a = out1['content']
fff.write(a.encode('utf8'))
fff.close()
script.write("python load_curator.py " + username + " " + pwd + " " + url + " schemas/" + title + str(counter) + " " + fn + " " + title + " ./schemas/" + title + str(counter) + "/files\n")
template_upload_url = "/rest/data/query/"
turl = url + template_upload_url
print 'status: Getting Data files associated with template..' + title + ":" +templ
data = {"query": "{}", "all": "true", "templates": "[{\"id\": \"" + templ + "\"}]"}
response2 = requests.post(turl, data, verify=False, auth=(username, pwd))
dout = response2.json()
return_code = subprocess.call("mkdir ./schemas/" + title + str(counter) + "/files", shell=True)
for dataf in dout:
print "--------------------------" + dataf['title']
fff = open("./schemas/" + title + str(counter) +"/files/" + dataf['title'], "w+")
a = dataf['xml_content']
a = a.replace(url,"http://127.0.0.1")
fff.write(a.encode('utf8'))
fff.close()
result = list1.findall(a)
for bid in result:
print "Result: " + str(result)
if bid:
blobid = save_blob("./schemas/" + title + str(counter) +"/blobs/", username, pwd,url,bid)
if (blobid != 0):
print "save: " + blobid
else:
print "Error could not open: " + bid + "!"
response_code = response.status_code
script.close()
if response_code == requests.codes.ok:
print "- status: Templates/files/blobs have been downloaded."
else:
response.raise_for_status()
raise Exception("error: a problem occurred when downloadong the schema (Error ", response_code, ")")
print 'status: done.'
def save_blob(dir='', username='',pwd='',url='',bid=''):
"""
Parameters: username - username on CDCS instance (Note: User must have admin privileges)
password - password on CDCS instance
url - url of CDCS instance
bid - id of blob, see if it exists, if not, store it and return new id
"""
print 'id: ' + str(bid)
if not os.path.exists(dir):
os.mkdir(dir)
xml_upload_url = "/rest/blob/" + bid + "/"
turl = url + xml_upload_url
response = requests.get(turl, verify=False, auth=(username, pwd), stream=True)
out = response.json()
xml_upload_url = "/rest/blob/download/" + bid + "/"
turl = url + xml_upload_url
blobfile = requests.get(turl, verify=False, auth=(username, pwd), stream=True)
fname = out['id'] + "_"+out['filename']
with open(dir + fname, 'wb') as fd:
fd.write(blobfile.content)
fd.close();
response_code = blobfile.status_code
if response_code == requests.codes.ok:
print "status: blob created: " + fname
ret = bid
else:
print "status: Error getting blob: " + response_code
ret = 0
return ret
OUTPUT:
status: getting Templates...http://vm-itl-ssd-017.nist.gov:8028/rest/template-version-manager/global
Working on;TracerDiffusivity Current: 5c8022a4d2d20554811857fc
200
status: Getting Data files associated with template..TracerDiffusivity:5c7801c5d2d2050ce2daf81c
200
status: Getting Data files associated with template..TracerDiffusivity:5c7801c8d2d2050ce3daf824
200
status: Getting Data files associated with template..TracerDiffusivity:5c8022a4d2d20554811857fc
--------------------------Al-Messer-1974
--------------------------Al-Beyeler1968.xml
--------------------------Al-Volin-1968.xml
--------------------------Ni-Bakker1968.xml
--------------------------Au-Impurity-Al-Peterson1970.xml
--------------------------AlFradin-1967.xml
--------------------------AlLundy1962.xml
--------------------------Al-Stoebe-1965.xml
--------------------------Al-Ni-Gust-1981.xml
--------------------------Al-Ti Araujo-2000.xml
Working on;MaterialDescription Current: 5c7801c8d2d2050ce4daf824
200
status: Getting Data files associated with template..MaterialDescription:5c7801c8d2d2050ce4daf824
Working on;Interdiffusion Current: 5c7f4ce5d2d20554801831c4
200
status: Getting Data files associated with template..Interdiffusion:5c7801c9d2d2050ce5daf824
200
status: Getting Data files associated with template..Interdiffusion:5c7801c9d2d2050ce2daf826
200
status: Getting Data files associated with template..Interdiffusion:5c7801cad2d2050ce5daf826
200
status: Getting Data files associated with template..Interdiffusion:5c7801cad2d2050ce4daf826
200
status: Getting Data files associated with template..Interdiffusion:5c7801cbd2d2050ce1daf828
200
status: Getting Data files associated with template..Interdiffusion:5c7801cbd2d2050ce3daf825
200
status: Getting Data files associated with template..Interdiffusion:5c7f34abd2d2055482184205
200
status: Getting Data files associated with template..Interdiffusion:5c7f38c5d2d2055483184429
200
status: Getting Data files associated with template..Interdiffusion:5c7f462bd2d20554831846e6
200
status: Getting Data files associated with template..Interdiffusion:5c7f4ce5d2d20554801831c4
--------------------------Obata-Co3Al-Co9Al3W-2014.xml
--------------------------GE-DiffusionCouple-IN718-R95.xml
Result: [u'5c7801ced2d2050ce2daf91f', u'5c7801cfd2d2050ce3daf839']
id: 5c7801ced2d2050ce2daf91f
status: blob created: 5c7801ced2d2050ce2daf91f_in718-r95-gray2.tif
save: 5c7801ced2d2050ce2daf91f
Result: [u'5c7801ced2d2050ce2daf91f', u'5c7801cfd2d2050ce3daf839']
id: 5c7801cfd2d2050ce3daf839
status: blob created: 5c7801cfd2d2050ce3daf839_IN718-R95.png
save: 5c7801cfd2d2050ce3daf839
--------------------------GE-DiffusionCouple-R95-R88.xml
Result: [u'5c7801d1d2d2050ce3daf927', u'5c7801d1d2d2050ce1daf832']
id: 5c7801d1d2d2050ce3daf927
status: blob created: 5c7801d1d2d2050ce3daf927_r95r88-1000h-labelled.tif
save: 5c7801d1d2d2050ce3daf927
Result: [u'5c7801d1d2d2050ce3daf927', u'5c7801d1d2d2050ce1daf832']
id: 5c7801d1d2d2050ce1daf832
status: blob created: 5c7801d1d2d2050ce1daf832_R95-R88-1000h.png
save: 5c7801d1d2d2050ce1daf832
--------------------------GE-DiffusionCouple-R88-IN100.xml
Result: [u'5c7801d2d2d2050ce3daf967', u'5c7801d2d2d2050ce1daf838']
id: 5c7801d2d2d2050ce3daf967
status: blob created: 5c7801d2d2d2050ce3daf967_R88-IN100-gray2.tif
save: 5c7801d2d2d2050ce3daf967
Result: [u'5c7801d2d2d2050ce3daf967', u'5c7801d2d2d2050ce1daf838']
id: 5c7801d2d2d2050ce1daf838
status: blob created: 5c7801d2d2d2050ce1daf838_R88-IN100.png
save: 5c7801d2d2d2050ce1daf838
--------------------------GE-DiffusionCouple-Ni-R88.xml
Result: [u'5c7801d3d2d2050ce3daf96e']
id: 5c7801d3d2d2050ce3daf96e
status: blob created: 5c7801d3d2d2050ce3daf96e_nir88-1000h-3.TIF
save: 5c7801d3d2d2050ce3daf96e
--------------------------TiV-TiAl_923K_2400h.xml
--------------------------Ti-TiV_923K_2400h.xml
--------------------------GE-DiffusionCouple-IN100-IN718.xml
Working on;TransitionTemperatures Current: 5c7801dad2d2050ce2daf94c
200
status: Getting Data files associated with template..TransitionTemperatures:5c7801d5d2d2050ce4daf827
200
status: Getting Data files associated with template..TransitionTemperatures:5c7801dad2d2050ce2daf94c
--------------------------Fe-19Ni-Ms-KaufmanCohen1956.xml
--------------------------Fe-28Ni-Ms-KaufmanCohen1956.xml
--------------------------Fe-24Ni-Ms-KaufmanCohen1956.xml
--------------------------Fe-29Ni-Ms-KaufmanCohen1956.xml
--------------------------Fe-30Ni-Ms-KaufmanCohen1956.xml
--------------------------Fe-14Ni-Ms-KaufmanCohen1956.xml
--------------------------CoAlW-L04.xml
--------------------------CoAlWbase-L19.xml
--------------------------CoAlW-L06.xml
--------------------------Co8Al8W4Cr2Ta-Suuzuki-ActaMater2008.xml
--------------------------CoAlW-L09.xml
--------------------------CoAlW-16Fe12B-Bauer2012.xml
--------------------------CoAlW-CrTa-Tsunekane-2011.xml
--------------------------CoAlWbase-L22.xml
--------------------------CoAlW-L05.xml
--------------------------CoAlWbase-L17.xml
--------------------------CoAlWbase-L16.xml
--------------------------CoAlWbase-L18.xml
--------------------------CoAlW-4B-Bauer2012.xml
--------------------------CoAlW-12B-Bauer2012.xml
--------------------------CoAlWbase-L19C.xml
--------------------------CoAlW-L08.xml
--------------------------CoAlW-Makineni2015.xml
--------------------------CoAlW-L02.xml
--------------------------CoAlW-L03.xml
--------------------------CoAlW-1Mo4B-Bauer2012.xml
--------------------------Co10Cr10Ni-OMori-2014.xml
--------------------------CoAlWNi-L11.xml
--------------------------CoAlWTa-Tsunekane-2011.xml
--------------------------CoAlW-Tsunekane-2011.xml
--------------------------CoAlWbase-L14.xml
--------------------------CoAlWbase-L13.xml
--------------------------CoAlWbase-L21.xml
--------------------------CoAlWNi-L10.xml
--------------------------Shinagawa-CoAlWNi-2008.xml
--------------------------CoAlWbase-L15.xml
--------------------------CoAlW-16Cr4B-Bauer2012.xml
--------------------------CoAlW-8Cr18Ni12B-Bauer2012.xml
--------------------------CoAlW-2Si-Bauer2012.xml
--------------------------CoAlWbase-L20.xml
--------------------------CoAlW-8Fe12B-Bauer2012.xml
--------------------------CoAlWNi-L07.xml
--------------------------CoAlW-2Ti12B-Bauer2012.xml
--------------------------CoAlWbase-L12.xml
--------------------------CoAlW-8Cr9Ni12B-Bauer2012.xml
Working on;ElectrochemicalPolarization Current: 5c7801ded2d2050ce1daf865
200
status: Getting Data files associated with template..ElectrochemicalPolarization:5c7801ddd2d2050ce5daf857
200
status: Getting Data files associated with template..ElectrochemicalPolarization:5c7801ddd2d2050ce3daf993
200
status: Getting Data files associated with template..ElectrochemicalPolarization:5c7801ded2d2050ce5daf859
200
status: Getting Data files associated with template..ElectrochemicalPolarization:5c7801ded2d2050ce1daf865
--------------------------AM17-4-PSU-HCl.xml
Result: [u'5c7801dfd2d2050ce2daf953']
id: 5c7801dfd2d2050ce2daf953
status: blob created: 5c7801dfd2d2050ce2daf953_20171129_AM17-HT-PSU-35HCl-PD2.xlsx
save: 5c7801dfd2d2050ce2daf953
- status: Templates/files/blobs have been downloaded.
status: done.
```python
def load_curator(username='',pwd='',url='',schema_path='',schema_filename='', schema_title='', xml_dir=''):
"""
load_curator: import a template and all data adn blob files into curator.
notes: reads a directory structure as so: (As created by dump_curator)
(Template Title)
(schema file.xsd)
(data files) (blob files)
(data_file1.xml) (blob_file1)
(data_file2.xml) (blob_file2)
(data_file3.xml) (blob_file3)
(data_file4.xml) (blob_file4)
(data_file5.xml) .
. .
. .
. .
(data_fileN.xml) (blob_fileX)
This strucure is created by dump_curator routine in this document. It will pre-pend the blobfile with its id used in the data file.
Parameters: username - username on CDCS instance (Note: User must have admin privileges)
password - password on CDCS instance
url - url of CDCS instance
schema_path - Directory path to XSD schema file
schema_filename - name of XSD schema file
schema_title - title to use for the Template in CDCS whne uploading XSD schema file
xml_dir - Directory path to XML Data files
"""
schema = join(schema_path, schema_filename) # When downloading, current schema is pre-pended with Cur_
if schema_filename[0:4] == 'Cur_':
is_current=1;
schema_filename = schema_filename[4:]
else:
is_current=0;
print "Schema Filename = " + schema_filename + " Iscurrent=" + str (is_current)
#
# If oid is pre-pended to filename remove it
#
oldid = schema_filename[0:24]
if all(c in string.hexdigits for c in oldid):
newfile = schema_filename[25:]
else:
newfile = schema_filename
with open(schema, "rb") as template_file:
template_content = template_file.read()
#
# First - see if template title already exists, add to it if it does, add newif it does not
#
template_upload_url = "/rest/template-version-manager/global/"
turl = url + template_upload_url
print 'status: checking for template to use when uploading xml files ...'
response = requests.get(turl, auth=(username, pwd))
response_content = json.loads(response.text)
print "Get:"
print turl
response_code = response.status_code
print "Resp: " + str(response_code)
#
# This will be default if we do not find it
#
template_upload_url = "/rest/template/global/"
data = {
"title": schema_title,
"filename": newfile,
"content": template_content
}
patch_url = ""
for rec in response_content:
if ( schema_title == rec['title']):
#
# Found it, use these instead
#
id=rec['id']
print "Found id=" + str(id)
template_upload_url = "/rest/template-version-manager/" + id + "/version/"
patch_url = url + "/rest/template/version/" + id + "/current/"
data = {
"filename": schema_filename,
"content": template_content
}
turl = url + template_upload_url
response = requests.post(turl, json=data, auth=(username, pwd))
print "Post: " + turl
response_content = json.loads(response.text)
if response_code == requests.codes.created:
template_id = str(response_content['id'])
print "status: Template has been uploaded with template_id = " + str (template_id)
if (is_current ==1):
patch_url = url + "/rest/template/version/" + template_id + "/current/"
print "patch: "+ patch_url
response = requests.patch(patch_url, auth=(username, pwd))
else:
raise Exception("- error: a problem occurred when uploading the schema (Error " + str(response_code) + ")")
# upload the directory full of XML files using:
# the XML schema : schema/(title).xsd
# the set of XML files: schema/(title)/files/(xml filenames)*.xml
# look in the data files for blobs that need loading
print 'Status: uploading xml files ...'
xml_upload_url = "/rest/data/"
for xml_filename in listdir(xml_dir):
with codecs.open(join(xml_dir, xml_filename), mode='r', encoding='utf-8') as xml_file:
xml_content = xml_file.read()
turl = url + xml_upload_url
oldid = xml_filename[0:24]
if all(c in string.hexdigits for c in oldid):
newfile = xml_filename[25:]
else:
newfile = xml_filename
xml_content =xml_content.replace('http://127.0.0.1', url)
list1 = re.compile(url + '\/rest\/blob\/download\/(.*?)\<')
result = list1.findall(xml_content)
print "Result"
print result
for bid in result:
if bid:
print "List1:" + bid + " file:" + xml_filename
blobid = load_blob(username, pwd,url,schema_path,bid)
if (blobid != 0):
xml_content = xml_content.replace(str(bid),str(blobid))
print "replace - " + bid + " -" + blobid
else:
print "Error could not open: " + bid + "!"
print url
dfile = upload_data(username, pwd,url,join(xml_dir, xml_filename),template_id)
if dfile == requests.codes.created:
print "status: " + xml_filename + " has been uploaded."
else:
print "Error: Upload failed with status code " + str(dfile)
print 'status: done.'
OUTPUT:
load: TracerDiffusivity uploaded.
Schema Filename = DTracerImpurity2.xsd Iscurrent=1
status: checking for template to use when uploading xml files ...
Get:
http://vm-itl-ssd-017.nist.gov:8090/rest/template-version-manager/global/
Resp: 200
Post: http://vm-itl-ssd-017.nist.gov:8090/rest/template/global/
Resp: 201
status: Template has been uploaded with template_id = 5c87f56ad2d205116e398a13
patch: http://vm-itl-ssd-017.nist.gov:8090/rest/template/version/5c87f56ad2d205116e398a13/current/
Status: uploading xml files ...
Result
[]
load: MaterialDescription uploaded.
Schema Filename = Material5.xsd Iscurrent=1
status: checking for template to use when uploading xml files ...
Get:
http://vm-itl-ssd-017.nist.gov:8090/rest/template-version-manager/global/
Resp: 200
Post: http://vm-itl-ssd-017.nist.gov:8090/rest/template/global/
Resp: 201
status: Template has been uploaded with template_id = 5c87f571d2d2051171398a1b
patch: http://vm-itl-ssd-017.nist.gov:8090/rest/template/version/5c87f571d2d2051171398a1b/current/
Status: uploading xml files ...
status: done.
load: Interdiffusion uploaded.
Schema Filename = Interdiffusion5.xsd Iscurrent=1
status: checking for template to use when uploading xml files ...
Get:
http://vm-itl-ssd-017.nist.gov:8090/rest/template-version-manager/global/
Resp: 200
Post: http://vm-itl-ssd-017.nist.gov:8090/rest/template/global/
Resp: 201
status: Template has been uploaded with template_id = 5c87f577d2d205116f398a1b
patch: http://vm-itl-ssd-017.nist.gov:8090/rest/template/version/5c87f577d2d205116f398a1b/current/
Status: uploading xml files ...
Result
[]
load: ElectrochemicalPolarization uploaded.
Schema Filename = 5a981c66974a230087d3a734_ElectroChemSchema.xsd Iscurrent=1
status: checking for template to use when uploading xml files ...
Get:
http://vm-itl-ssd-017.nist.gov:8090/rest/template-version-manager/global/
Resp: 200
Post: http://vm-itl-ssd-017.nist.gov:8090/rest/template/global/
Resp: 201
status: Template has been uploaded with template_id = 5c87f598d2d205116d398a43
patch: http://vm-itl-ssd-017.nist.gov:8090/rest/template/version/5c87f598d2d205116d398a43/current/
Status: uploading xml files ...
Result
[u'5c7801dfd2d2050ce2daf953']
List1:5c7801dfd2d2050ce2daf953 file:AM17-4-PSU-HCl.xml
id: 5c7801dfd2d2050ce2daf953
Blobfile: schemas/ElectrochemicalPolarization4/blobs/5c7801dfd2d2050ce2daf953_20171129_AM17-HT-PSU-35HCl-PD2.xlsx
New: 20171129_AM17-HT-PSU-35HCl-PD2.xlsx
Check Blob:http://vm-itl-ssd-017.nist.gov:8090/rest/blob/5c7801dfd2d2050ce2daf953/
{u'message': u'Blob not found.'}
Get:
Resp: 404
Open blob
Open New
http://vm-itl-ssd-017.nist.gov:8090
Post:
json:
201
{u'handle': u'http://vm-itl-ssd-017.nist.gov/rest/blob/download/5c87f599d2d2051170398a4b/', u'upload_date': u'2019-03-12 18:08:25+00:00', u'user_id'
: u'2', u'id': u'5c87f599d2d2051170398a4b', u'filename': u'20171129_AM17-HT-PSU-35HCl-PD2.xlsx'}
Resp:
(Response [201])
status: blob uploaded.
Return: 5c87f599d2d2051170398a4b
replace - 5c7801dfd2d2050ce2daf953 -5c87f599d2d2051170398a4b
http://vm-itl-ssd-017.nist.gov:8090
status: uploading data file...
Filename:AM17-4-PSU-HCl.xml
Template_id:5c87f598d2d205116d398a43
status: AM17-4-PSU-HCl.xml has been uploaded.
status: done.
#Utilities
(xslt DIR)
(xslt1)
(xslt2)
(xslt3)
(xslt4)
(xslt5)
.
.
(xsltn)
This example use - xslts (directory where xslt files are)
def upload_xslt(username='',pwd='',url='',xml_dir=''):
"""
Parameters: username - username on CDCS instance (Note: User must have admin provledges)
password - password on CDCS instance
url - url of CDCS instance
xml_dir - Directory path to XSLT Data files
"""
print '- status: uploading xslt files ...'
xml_upload_url = "/rest/xslt/"
for xml_filename in listdir(xml_dir):
with open(join(xml_dir, xml_filename), "r") as xml_file:
xml_content = xml_file.read()
turl = url + xml_upload_url
print url
print xml_filename
data = {
"name": xml_filename,
"filename": xml_filename,
"instance_name": xml_filename,
"title": xml_filename,
"content": xml_content
}
print "Post:"
print turl
print "turl:"
response_str = requests.post(turl, data, auth=(username, pwd))
response_code = response_str.status_code
print "Resp: "
#print response_str.text
response = json.loads(response_str.text)
if response_code == HTTP_201_CREATED:
print "- status: uploaded."
else:
print "- error: Upload failed with status code "
print response_code
print ":"
print response["message"]
##OUTPUT
This example use - xslts (directory where xslt files are)
- status: uploading xslt files ...
http://vm-itl-ssd-017.nist.gov:8090
export_lib.xslt
Post:
http://vm-itl-ssd-017.nist.gov:8090/rest/xslt/
turl:
Resp:
- status: uploaded.
http://vm-itl-ssd-017.nist.gov:8090
export_map.xslt
Post:
http://vm-itl-ssd-017.nist.gov:8090/rest/xslt/
turl:
Resp:
- status: uploaded.
http://vm-itl-ssd-017.nist.gov:8090
export_spectra.xslt
Post:
http://vm-itl-ssd-017.nist.gov:8090/rest/xslt/
turl:
Resp:
- status: uploaded.
http://vm-itl-ssd-017.nist.gov:8090
export_xrd.xslt
Post:
http://vm-itl-ssd-017.nist.gov:8090/rest/xslt/
turl:
Resp:
- status: uploaded.
This example uses input template title - Interdiffusion
def export_data_by_template(username='',pwd='',url='', template_file_title=''):
"""
export_data_by_template: script to export data from a curator by template in JSON format
Parameters: username - username on CDCS instance (Note: User must have admin privileges)
password - password on CDCS instance
url - url of CDCS instance
template_file_title - title of Template in CDCS to get all data files uploaded using it.
"""
template_url = "/rest/template-version-manager/global"
turl = url + template_url
current=''
print 'status: getting Template... Title:' + template_file_title
response = requests.get(turl, verify=False, auth=(username, pwd))
out = json.loads(response.text)
for rec in out:
if rec['title'] == template_file_title:
current=rec['current']
print "Found:" + current
query_url = "/rest/data/query/"
turl = url + query_url
if current == '':
response.raise_for_status()
raise Exception("Error: No Template named (" + template_file_title + ")")
id_list = []
print 'status: Query Data using template...'
data = {"query": "{}", "all": "true", "templates": "[{\"id\":\"" + current + "\"}]"}
print "Post:"
response= requests.post(turl, data=data, auth=(username, pwd))
out = response.json()
print "Resp: "
print response.status_code
response_code = response.status_code
ids = [did['id'] for did in out]
print "ID:" + str(ids)
# get JSON Exporter
exporter_url = "/exporter/rest/exporter/"
turl = url + exporter_url
response = requests.get(turl, verify=False, auth=(username, pwd))
out = json.loads(response.text)
exporter_id = []
for rec in out:
if rec['name'] == "JSON":
exporter_id.append(rec['id'])
print "Found JSON exporter:"
exporter_url = "/exporter/rest/exporter/export/"
turl = url + exporter_url
print 'status: Exporting Data files using template...'
data = {"exporter_id_list":exporter_id,"data_id_list": ids}
print "Post:"
response= requests.post(turl, json=data, auth=(username, pwd))
out = response.json()
pprint(out)
print "Resp: "
print response.status_code
response_code = response.status_code
if response_code == requests.codes.ok:
print "status: downloaded."
else:
response.raise_for_status()
raise Exception("Error: a problem occurred when uploading the schema (Error ", response_code, ")")
print '- status: done.'
# get zip file
exporter_url = "/exporter/rest/exporter/export/download/"+ out['id'] + "/"
turl = url + exporter_url
print turl
blobfile = requests.get(turl, verify=False, auth=(username, pwd), stream=True)
response_code = blobfile.status_code
print 'Unziping.'
return_code = subprocess.call("rm -rf ./JSON_Files", shell=True)
return_code = subprocess.call("mkdir ./JSON_Files", shell=True)
zip = zipfile.ZipFile(StringIO.StringIO(blobfile.content))
zip.extractall('./JSON_Files')
if response_code == requests.codes.ok:
print "status: JSON Files created: "
else:
response.raise_for_status()
raise Exception("Error getting data (Error ", response_code, ")")
##OUTPUT
This example uses input template title - Interdiffusion
status: getting Template... Title:Interdiffusion
Found:5c7d88edd2d2052570d4e8aa
status: Query Data using template...
Post:
Resp:
200
ID:[u'5c7d88f1d2d205256cd4e8ab', u'5c7d88f4d2d205256cd4e8af', u'5c7d88f6d2d205256dd4e8a9', u'5c7d88f7d2d205256dd4e8ad', u'5c7d88fcd2d205256dd4e8b1', u'5c7d88fdd2d2052570d4e8c1', u'5c7d88fdd2d205256cd4e9e2', u'5c7d88fed2d205256ed4e8ab']
Found JSON exporter:
status: Exporting Data files using template...
Post:
{u'id': u'5c7fdf77d2d205256fd4ea13'}
Resp:
200
status: downloaded.
- status: done.
http://vm-itl-ssd-017.nist.gov:8090/exporter/rest/exporter/export/download/5c7fdf77d2d205256fd4ea13/
Unziping.
Select all templates from a 1.X CDCS instance and write to a JSON file. You will use this to extract all templates out of your 1.X instance of MDCS and write it to a JSON file. You use this file later to import your 1.X data into a 2.X instance.
#!/usr/bin/python
program: get_template.py
description:
- script to extract templates from a 1.X MDCS and write all templates to a json file for later upload to a 2.X instance.
notes:
- to customize for a given mdcs instance, change the variable settings below:
- user_name
- user_pwd
- mdcs URL
from os.path import join
from os import listdir
import requests
import json
import sys
import os
def upload_xml_for_schema(username='',pwd='',url=''):
# MDCS username / password
# username = "(username)"
# pwd = "(password)"
template_upload_url = "/rest/templates/select/all"
turl = url + template_upload_url
# upload the schema using REST
print 'status: getting Templates...'
print "Get:"
response = requests.get(turl, verify=False, auth=(username, pwd))
response.json()
with open('Template.json', 'wb') as fd:
for chunk in response.iter_lines(chunk_size=512, decode_unicode=None, delimiter=None):
fd.write(chunk)
response_code = response.status_code
print "Response Code: "
print response.status_code
if response_code == requests.codes.ok:
print "status: Template.json has been downloaded."
else:
response.raise_for_status()
print 'status: done.'
def main(argv=[]):
if ( len(argv) != 4 ):
print 'usage: python get_template.py (username) (password) (url) '
print 'example: python get_template.py blong my_password http://127.0.0.1:8000 '
exit
args=argv[1:] # skip arg[0]=bulk_upload.py
username=args[0]
pwd=args[1]
url=args[2]
upload_xml_for_schema(username,pwd,url)
if (__name__ == '__main__' ):
main(sys.argv)
Select all data files from a 1.X CDCS instance and write to a JSON file. You will use this to extract all data out of your 1.X instance of MDCS and write it to a JSON file. You use this file later to import your 1.X data into a 2.X instance.
#!/usr/bin/python
"""
program: get_data.py
description:
- script to extract data from a 1.X MDCS and write all data to a json file for later upload to a 2.X instance.
notes:
- to customize for a given mdcs instance, change the variable settings below:
- user_name
- user_pwd
- mdcs URL
"""
from os.path import join
from os import listdir
import requests
import json
import sys
import os
def upload_xml_for_schema(username='',pwd='',url=''):
# MDCS username / password
# username = "(username)"
# pwd = "(password)"
template_upload_url = "/rest/explore/select/all"
turl = url + template_upload_url
# get data using REST
print 'status: Getting Data file...'
print "Get:"
response = requests.get(turl, verify=False, auth=(username, pwd),stream=True)
with open('Data.json', 'wb') as fd:
for chunk in response.iter_content(chunk_size=512):
fd.write(chunk)
response_code = response.status_code
print "Response Code: "
print response.status_code
if response_code == requests.codes.ok:
print "status: Data.json downloaded."
else:
response.raise_for_status()
print 'status: done.'
def main(argv=[]):
if ( len(argv) != 4 ):
print 'usage: python get_data.py (username) (password) (url) '
print 'example: python get_data.py blong my_password http://127.0.0.1:8000 '
exit
args=argv[1:] # skip arg[0]=bulk_upload.py
username=args[0]
pwd=args[1]
url=args[2]
upload_xml_for_schema(username,pwd,url)
if (__name__ == '__main__' ):
main(sys.argv)
Select all binary data files from a 1.X CDCS instance and write to a JSON file. You will use this to extract all binary data out of your 1.X instance of MDCS and write it to a JSON file. You use this file later to import your 1.X binary data into a 2.X instance.
#!/usr/bin/python
"""
program: get_blob.py
description:
- script to export all blob data from a 1.X curator to Blob file for later import into a 2.X instance.
notes:
- to customize for a given mdcs instance, change the variable settings below:
- user_name
- user_pwd
- url
"""
from os.path import join
from os import listdir
import requests
import json
import sys
import os
def download_blob_data(username='',pwd='',url=''):
# MDCS username / password
# username = "(username)"
# pwd = "(password)"
blob_upload_url = "/rest/blob/list"
turl = url + blob_upload_url
# get blob data using REST
print 'status: Getting Blob files...'
print "Get:"
response = requests.get(turl, verify=False, auth=(username, pwd))
with open('Blob.json', 'wb') as fd:
for chunk in response.iter_lines(chunk_size=512, decode_unicode=None, delimiter=None):
fd.write(chunk)
response_code = response.status_code
print "Response Code: "
print response.status_code
if response_code == requests.codes.ok:
print "status: Blob.json has been downloaded."
else:
response.raise_for_status()
raise Exception("- error: a problem occurred when downloading the blobs (Error ", response_code, ")")
print 'status: done.'
def main(argv=[]):
if ( len(argv) != 4 ):
print 'usage: python get_blob.py (username) (password) (url) '
print 'example: python get_blob.py blong my_password http://127.0.0.1:8000 '
exit
args=argv[1:] # skip arg[0]=get_blob.py
username=args[0]
pwd=args[1]
url=args[2]
download_blob_data(username,pwd,url)
if (__name__ == '__main__' ):
main(sys.argv)